Research

Publications
- Neri I, Ramazzotti G, Mongiorgi S, Rusciano I, Bugiani M, Conti L, Cousin M, Giorgio E, Padiath QS, Vaula G, Cortelli P, Manzoli L, Ratti S. Understanding the Ultra-Rare Disease Autosomal Dominant Leukodystrophy: an Updated Review on Morpho-Functional Alterations Found in Experimental Models. Mol. Neurobiol. 2023. doi: 10.1007/s12035-023-03461-1.
- Evangelisti C, Rusciano I, Mongiorgi S, Ramazzotti G, Lattanzi G, Manzoli L, Cocco L, Ratti S. The wide and growing range of lamin B-related diseases: from laminopathies to cancer. Cell Mol Life Sci. 2022 Feb 8;79(2):126. doi: 10.1007/s00018-021-04084-2. PMID: 35132494; PMCID: PMC8821503.
- Ratti S, Rusciano I, Mongiorgi S, Neri I, Cappellini A, Cortelli P, Suh PG, McCubrey JA, Manzoli L, Cocco L, Ramazzotti G. Lamin B1 Accumulation’s Effects on Autosomal Dominant Leukodystrophy (ADLD): Induction of Reactivity in the Astrocytes. Cells. 2021 Sep 28;10(10):2566. doi: 10.3390/cells10102566. PMID: 34685544; PMCID: PMC8534128.
- Ratti S, Rusciano I, Mongiorgi S, Owusu Obeng E, Cappellini A, Teti G, Falconi M, Talozzi L, Capellari S, Bartoletti-Stella A, Guaraldi P, Cortelli P, Suh PG, Cocco L, Manzoli L, Ramazzotti G. Cell signaling pathways in autosomal-dominant leukodystrophy (ADLD): the intriguing role of the astrocytes. Cell Mol Life Sci. 2021 Mar;78(6):2781-2795. doi: 10.1007/s00018-020-03661-1. Epub 2020 Oct 9. PMID: 33034697; PMCID: PMC8004488.
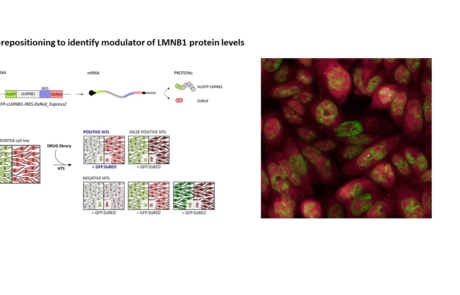
- Giorgio E, Pesce E, Pozzi E, Sondo E, Ferrero M, Morerio C, Borrelli G, Della Sala E, Lorenzati M, Cortelli P, Buffo A, Pedemonte N, Brusco A. A high-content drug screening strategy to identify protein level modulators for genetic diseases: A proof-of-principle in autosomal dominant leukodystrophy. Hum Mutat. 2021 Jan;42(1):102-116. doi: 10.1002/humu.24147. Epub 2020 Dec 8. PMID: 33252173.
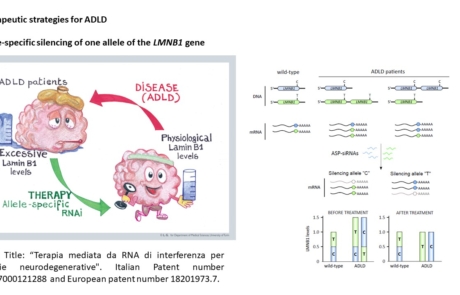
- Giorgio E, Lorenzati M, Rivetti di Val Cervo P, Brussino A, Cernigoj M, Della Sala E, Bartoletti Stella A, Ferrero M, Caiazzo M, Capellari S, Cortelli P, Conti L, Cattaneo E, Buffo A, Brusco A. Allele-specific silencing as treatment for gene duplication disorders: proof-of-principle in autosomal dominant leukodystrophy. Brain. 2019 Jul 1;142(7):1905-1920. doi: 10.1093/brain/awz139. PMID: 31143934.
- Nmezi B, Giorgio E, Raininko R, Lehman A, Spielmann M, Koenig MK, Adejumo R, Knight M, Gavrilova R, Alturkustani M, Sharma M, Hammond R, Gahl WA, Toro C, Brusco A, Padiath QS. Genomic deletions upstream of lamin B1 lead to atypical autosomal dominant leukodystrophy. Neurol Genet. 2019 Jan 24;5(1):e305. doi: 10.1212/NXG.0000000000000305. Erratum in: Neurol Genet. 2019 Sep 23;5(5):e362. PMID: 30842973; PMCID: PMC6384018.
- Lo Martire V, Alvente S, Bastianini S, Berteotti C, Bombardi C, Calandra-Buonaura G, Capellari S, Cohen G, Cortelli P, Gasparini L, Padiath Q, Valli A, Zoccoli G, Silvani A. Mice overexpressing lamin B1 in oligodendrocytes recapitulate the age-dependent motor signs, but not the early autonomic cardiovascular dysfunction of autosomal-dominant leukodystrophy (ADLD). Exp Neurol. 2018 Mar;301(Pt A):1-12. doi: 10.1016/j.expneurol.2017.12.006. Epub 2017 Dec 17. PMID: 29262292; PMCID: PMC5809293.
- Terlizzi R, Calandra-Buonaura G, Zanigni S, Barletta G, Capellari S, Guaraldi P, Donadio V, Cason E, Contin M, Poda R, Tonon C, Sambati L, Gallassi R, Liguori R, Lodi R, Cortelli P. A longitudinal study of a family with adult-onset autosomal dominant leukodystrophy: Clinical, autonomic and neuropsychological findings. Auton Neurosci. 2016 Feb;195:20-6. doi: 10.1016/j.autneu.2016.02.005. Epub 2016 Feb 8. PMID: 26896090.
- Zanigni S, Terlizzi R, Tonon C, Testa C, Manners DN, Capellari S, Gallassi R, Poda R, Gramegna LL, Calandra-Buonaura G, Sambati L, Cortelli P, Lodi R. Brain magnetic resonance metabolic and microstructural changes in adult-onset autosomal dominant leukodystrophy. Brain Res Bull. 2015 Aug;117:24-31. doi: 10.1016/j.brainresbull.2015.07.002. Epub 2015 Jul 17. PMID: 26189928.
- Giorgio E, Robyr D, Spielmann M, Ferrero E, Di Gregorio E, Imperiale D, Vaula G, Stamoulis G, Santoni F, Atzori C, Gasparini L, Ferrera D, Canale C, Guipponi M, Pennacchio LA, Antonarakis SE, Brussino A, Brusco A. A large genomic deletion leads to enhancer adoption by the lamin B1 gene: a second path to autosomal dominant adult-onset demyelinating leukodystrophy (ADLD). Hum Mol Genet. 2015 Jun 1;24(11):3143-54. doi: 10.1093/hmg/ddv065. Epub 2015 Feb 20. PMID: 25701871; PMCID: PMC4424952.
- Bartoletti-Stella A, Gasparini L, Giacomini C, Corrado P, Terlizzi R, Giorgio E, Magini P, Seri M, Baruzzi A, Parchi P, Brusco A, Cortelli P, Capellari S. Messenger RNA processing is altered in autosomal dominant leukodystrophy. Hum Mol Genet. 2015 May 15;24(10):2746-56. doi: 10.1093/hmg/ddv034. Epub 2015 Jan 30. Erratum in: Hum Mol Genet. 2017 Oct 1;26(19):3868. PMID: 25637521; PMCID: PMC4406291.
- Ferrera D, Canale C, Marotta R, Mazzaro N, Gritti M, Mazzanti M, Capellari S, Cortelli P, Gasparini L. Lamin B1 overexpression increases nuclear rigidity in autosomal dominant leukodystrophy fibroblasts. FASEB J. 2014 Sep;28(9):3906-18. doi: 10.1096/fj.13-247635. Epub 2014 May 22. PMID: 24858279; PMCID: PMC4139899.
- Columbaro M, Mattioli E, Maraldi NM, Ortolani M, Gasparini L, D’Apice MR, Postorivo D, Nardone AM, Avnet S, Cortelli P, Liguori R, Lattanzi G. Oct-1 recruitment to the nuclear envelope in adult-onset autosomal dominant leukodystrophy. Biochim Biophys Acta. 2013 Mar;1832(3):411-20. doi: 10.1016/j.bbadis.2012.12.006. Epub 2012 Dec 20. Erratum in: Biochim Biophys Acta. 2013 Dec;1832(12):2244. PMID: 23261988.
- Giorgio E, Rolyan H, Kropp L, Chakka AB, Yatsenko S, Di Gregorio E, Lacerenza D, Vaula G, Talarico F, Mandich P, Toro C, Pierre EE, Labauge P, Capellari S, Cortelli P, Vairo FP, Miguel D, Stubbolo D, Marques LC, Gahl W, Boespflug-Tanguy O, Melberg A, Hassin-Baer S, Cohen OS, Pjontek R, Grau A, Klopstock T, Fogel B, Meijer I, Rouleau G, Bouchard JP, Ganapathiraju M, Vanderver A, Dahl N, Hobson G, Brusco A, Brussino A, Padiath QS. Analysis of LMNB1 duplications in autosomal dominant leukodystrophy provides insights into duplication mechanisms and allele-specific expression. Hum Mutat. 2013 Aug;34(8):1160-71. doi: 10.1002/humu.22348. Epub 2013 May 28. Erratum in: Hum Mutat. 2014 Jan;35(1):149. PMID: 23649844; PMCID: PMC3714349.
- Cortelli P, Terlizzi R, Capellari S, Benarroch E. Nuclear lamins: functions and clinical implications. Neurology. 2012 Oct 16;79(16):1726-31. doi: 10.1212/WNL.0b013e31826ea887. PMID: 23071165.
- Guaraldi P, Donadio V, Capellari S, Contin M, Casadio MC, Montagna P, Liguori R, Cortelli P. Isolated noradrenergic failure in adult-onset autosomal dominant leukodystrophy. Auton Neurosci. 2011 Jan 20;159(1-2):123-6. doi: 10.1016/j.autneu.2010.07.011. Epub 2010 Aug 16. PMID: 20719577.
- Brussino A, Vaula G, Cagnoli C, Panza E, Seri M, Di Gregorio E, Scappaticci S, Camanini S, Daniele D, Bradac GB, Pinessi L, Cavalieri S, Grosso E, Migone N, Brusco A. A family with autosomal dominant leukodystrophy linked to 5q23.2-q23.3 without lamin B1 mutations. Eur J Neurol. 2010 Apr;17(4):541-9. doi: 10.1111/j.1468-1331.2009.02844.x. Epub 2009 Dec 4. PMID: 19961535.
- Brussino A, D’Alfonso S, Cagnoli C, Di Gregorio E, Barberis M, Padovan S, Vaula G, Pinessi L, Squadrone S, Abete MC, Collimedaglia L, Guerini FR, Migone N, Brusco A. Mutations in the lamin B1 gene are not present in multiple sclerosis. Eur J Neurol. 2009 Apr;16(4):544-6. doi: 10.1111/j.1468-1331.2009.02536.x. PMID: 19348623.
Grants
Finalizzata GR-2021-12373348
Budget: 450,000 euro
Project duration: 3 years
Principal Investigator: Elisa Giorgio (Fondazione Istituto Neurologico Casimiro Mondino; Università di Pavia)
Co-PI: Stefano Ratti (Alma Mater Studiorum – Università di Bologna) e Emanuela Pesce (IRCCS Istituto Giannina Gaslini)
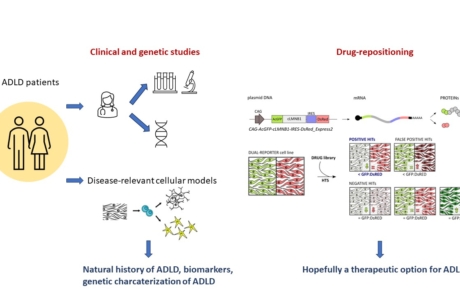
At present, there is no effective therapy to cure or slow the advance of Autosomal Dominant adult-onset demyelinating leukodystrophy (ADLD), a slowly progressive disease leading to demyelination caused by Lamin B1 (LMNB1) overexpression due to coding duplications or noncoding deletions at the LMNB1 locus. This project will i) pave the way towards a therapy for ADLD by “repositioning” FDA-approved agents able to restore physiological LMNB1 protein levels; ii) redefine ADLD pathology in patients and disease-relevant cellular models, and iii) contribute to a better understanding of the genotype-phenotype correlation of structural variants at the LMNB1 locus, allowing a better family counseling and patient care. In our best scenario, the proposed drug-repositioning might even lead to available drugs for patients following the “compassionate use” practice, strongly shortening the way to the cure.
EXPECTED OUTCOMES: Overall, our project will pave the way for developing specific therapies for ADLD and will generate human ADLD-relevant cellular models, strongly improving the knowledge on ADLD physiopathology. Moreover, we will provide a deeper understanding of the neuropathological course, define a panel of biomarkers to follow disease progression and to monitor the therapeutic efficacy of our strategies in future clinical trials. Finally, we will decipher the role of position effects in the 3D genome and TAD alterations as a new pathomechanism for ADLD, contributing to a better understanding of the genotype- phenotype correlation of structural variants at the LMNB1 locus. As conventional phenotyping and expression analysis of patient cells in neurodevelopmental phenotypes is challenging, the potential of 3C technologies in this project could provide a first step for the use of 3C techniques in a diagnostic context.
ADLD Center Grant
Project duration: 1 year
Budget: 50.000$
PI: Stefano Ratti (Alma Mater Studiorum – Università di Bologna)
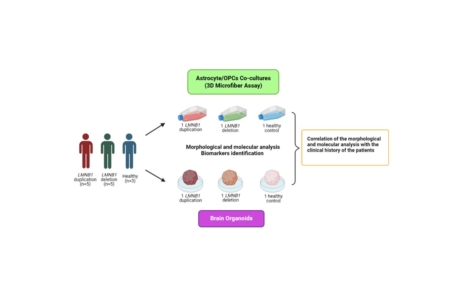
The project aims at developing reliable ADLD microfiber and organoid models for investigating biomarkers and for drug testing. The novel models to be developed with this substantial 1-year funding include 3D microfiber co-cultures of astrocytes and oligodendrocyte precursors (OPCs) and brain organoids. These models will be created from the fibroblasts of patients with the LMNB1 gene duplication and deletion phenotypes and healthy donors. The hiPSCs and NPCs needed for developing these models have already been produced and only differentiation of the NPCs to astrocytes and OPCs is required. Morphological and molecular analyses will take place for all the models to elucidate the unknown mechanisms of demyelination associated with Lamin B1. Moreover, studying specifically the synergic role of reactive astrocytes and oligodendrocytes in demyelination through the 3D co-culture models will be useful for testing drugs that target specific cellular populations. Furthermore, upgrading the research with innovative ADLD brain organoids will contribute to surpassing the difficulties faced with animal models and establishing brain-like reliable models for drug testing. Lastly, correlating the experimental outcomes of this study with the clinical history of the patients will be essential for establishing new treatments. In conclusion, the innovative 3D ADLD models to be created in this extensive 1-year investigation will provide new treatment directions for this rare and fatal disorder that lacks an effective therapy.